- HOME
- RESEARCH
- Research Units
- RNA Biology
RESEARCH
RNA Biology
Recent advances in RNA/DNA sequencing and genome-wide microarray technologies have revealed that transcription is widespread throughout the eukaryotic genomes, generating a large number of non-coding RNAs (ncRNAs). NcRNAs are defined as RNAs with no significant protein-coding capacity. Expression of ncRNAs is in some cases regulated developmentally and tissue-specifically. We are analyzing biological roles of ncRNAs in a wide range of cellular reactions, such as pre-mRNA splicing, nuclear structure formation, chromatin remodeling and genome stability, using fission yeast and cultured mammalian cells. NcRNAs can be categorized into two groups; short ncRNA, such as miRNAs and piRNAs, typically 20-200 bases in length, and long ncRNAs (lncRNA), such as satellite lncRNAs, ranging from ~200 bases to 100 kb in length. We have recently revealed that satellite lncRNAs transcribed from the centromere regions have essential roles in chromosome segregation, and suggested that these lncRNAs are promising targets for anti-cancer drugs. A new paradigm of “ncRNA regulation” is now emerging.
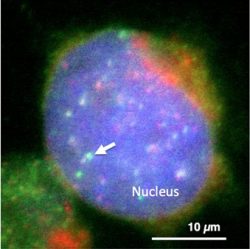
Figure legend: Visualization of satellite I lncRNA in the nucleus of human cultured cell. Satellite I lncRNA was found to be enriched at the centromere (Green: satellite I lncRNA, Red: centromere, Blue: DNA).
Unit members
-
 Unit coordinatorTokio TANI Website
Unit coordinatorTokio TANI WebsiteProfessor
Faculty of Advanced Science and Technology, Kumamoto UniversityJapan -
 Ramesh Shanmughom PILLAI Website
Ramesh Shanmughom PILLAI WebsiteProfessor
Department of Molecular Biology, University of GenevaSwitzerland -
 Takashi IDEUE Website
Takashi IDEUE WebsiteAssistant Professor
Faculty of Advanced Science and Technology, Kumamoto UniversityJapan
Achievements
Publications
Grants
KAKENHI and other external grants
- KAKENHI: Grant-in-Aid for Scientific Research (B) (3,200,000 yen)
- Other external grants: HIGIN Gap Grant (3,000,000 yen)
Patents
- Acquisition Patent 6744926: Novel plant growth inhibitor Kumamonamide
Activities
- Units of World-leading Researchers
-
- Development of Nano and Supramolecular Materials
- RNA Biology
- Plant Cell and Developmental Biology
- Nano-Organics and Nano-Hybrids
- Nano-medicine and Drug Delivery System
- Nano-medicine and Theranostics
- Multiscale Modeling of Soil and Rock Materials Using X-ray CT
- Quantification of Three Dimensional Vascular Network
- MicroCT-based Quantification of Fibrosis and Vascularization in Pancreatic Tumor
- Advanced Structural Materials
- Microstructure Analysis and Grain Boundary Engineering
- Structure and Dynamics of Materials Using Quantum Beams and Data-Driven Sciences
- Hydrological Environments
- Nano-materials for Energy Applications and Environmental Protection
- Units of Young Researchers
-
- Quantitative Bioimaging
- Development of Novel Therapeutic Strategy Using Iron Targeted Upconversion Nanoparticles for Parkinson's Disease
- Deep Learning for Hydrology
- Environmental Impacts of Ionic Solutes
- Study of first-generation objects in the universe with radio telescopes
- Plant Stem Cells and Regeneration
- Development of Microbially-Aided Carbon Sequestration Technology
- Advanced Biomedical Evaluation System
- Bio-inspired Functional Molecular System
- Nanomaterials Processing for Medical, Cosmetic, and Environmental Applications
- Ferroelectric Photovoltaics
- Next-Generation Design of Structures



